Challenge Open Access
Classification of Heart Sound Recordings: The PhysioNet/Computing in Cardiology Challenge 2016
Chengyu Liu , David Springer , Benjamin Moody , Ikaro Silva , Alistair Johnson , Maryam Samieinasab , Reza Sameni , Roger Mark , Gari D. Clifford
Published: March 4, 2016. Version: 1.0.0
Special issue of Physiological Measurement on "Recent advances in heart sound analysis" (Nov. 21, 2016, 12:42 p.m.)
The journal Physiological Measurement is hosting a special issue on “Recent advances in heart sound analysis”. We encourage all Challenge2016 entrants (and those who missed the opportunity to compete or attend CinC 2016) to submit extended analyses and articles to that issue, taking into account the publications and discussions at CinC 2016.
2016 CinC/PhysioNet Challenge papers (Nov. 21, 2016, 12:38 p.m.)
Papers and corresponding source code from the 2016 Challenge are now online.
More news
Sample code released for the PhysioNet/CinC Challenge 2016 (March 15, 2016, 1:44 p.m.)
Example code for Matlab and Octave has been released.
PhysioNet/CinC Challenge 2016 (March 4, 2016, 2 a.m.)
We are pleased to announce the 2016 PhysioNet/Computing in Cardiology Challenge: Classification of Normal/Abnormal Heart Sound Recordings. For this year's Challenge, we have released a collection of 3,125 phonocardiograms from a variety of clinical and non-clinical sources. We invite participants to develop algorithms to determine, based on a short heart sound recording, whether the patient should be referred on for an expert diagnosis.
Community forum for the PhysioNet/CinC Challenge 2016 (March 1, 2016, 12:48 p.m.)
If you have any questions or comments regarding this challenge, please post it directly in our Community Discussion Forum. This will increase transparency (benefiting all the competitors) and ensure that all the challenge organizers see your question.
Please include the standard citation for PhysioNet:
(show more options)
Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov, P. C., Mark, R., ... & Stanley, H. E. (2000). PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 101 (23), pp. e215–e220.
Introduction
The 2016 PhysioNet/CinC Challenge aims to encourage the development of algorithms to classify heart sound recordings collected from a variety of clinical or nonclinical (such as in-home visits) environments. The aim is to identify, from a single short recording (10-60s) from a single precordial location, whether the subject of the recording should be referred on for an expert diagnosis.
During the cardiac cycle, the heart firstly generates the electrical activity and then the electrical activity causes atrial and ventricular contractions. This in turn forces blood between the chambers of the heart and around the body. The opening and closure of the heart valves is associated with accelerations-decelerations of blood, giving rise to vibrations of the entire cardiac structure (the heart sounds and murmurs) [1]. These vibrations are audible at the chest wall, and listening for specific heart sounds can give an indication of the health of the heart. The phonocardiogram (PCG) is the graphical representation of a heart sound recording. Figure 1 illustrates a short section of a PCG recording.
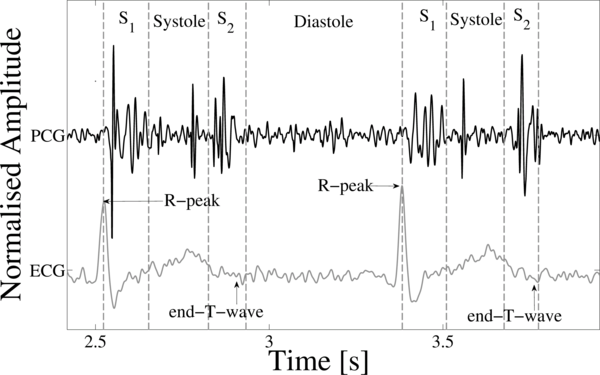
Figure 1. A PCG (center tracing), with simultaneously recorded ECG (lower tracing) and the four states of the PCG recording; S1, Systole, S2 and Diastole.
Four locations are most often used to listen to the heart sounds, which are named according to the positions where the valves can be best heard:
- Aortic area - centered at the second right intercostal space.
- Pulmonic area - in the second intercostal space along the left sternal border.
- Tricuspid area - in the fourth intercostal space along the left sternal edge.
- Mitral area - at the cardiac apex, in the fifth intercostal space on the midclavicular line.
Fundamental heart sounds (FHSs) usually include the first (S1) and second (S2) heart sounds. S1 occurs at the beginning of isovolumetric ventricular contraction, when the mitral and tricuspid valves close due to the rapid increase in pressure within the ventricles. S2 occurs at the beginning of diastole with the closure of the aortic and pulmonic valves. While the FHSs are the most recognizable sounds of the heart cycle, the mechanical activity of the heart may also cause other audible sounds, such as the third heart sound (S3), the fourth heart sound (S4), systolic ejection click (EC), mid-systolic click (MC), diastolic sound or opening snap (OS), as well as heart murmurs caused by the turbulent, high-velocity flow of blood.
The segmentation of the FHSs is a first step in the automatic analysis of heart sounds. The accurate localization of the FHSs is a prerequisite for the identification of the systolic or diastolic regions, allowing the subsequent classification of pathological situations in these regions [2]. Challenge participants could refer to the literature [3-10] for a quick review of previously developed segmentation methods.
The automated classification of pathology in heart sound recordings has been performed for over 50 years, but still presents challenges. Gerbarg et al were the first researchers to attempt the automatic classification of pathology in PCGs using a threshold-based method [11], motivated by the need to identify children with rheumatic heart disease (RHD). Artificial neural networks (ANNs) have been the most widely used machine learning-based approach for heart sound classification. Typical relevant studies grouped by the signal features as the input to the ANN classifier include: using wavelet features [12], time, frequency and complexity-based features [13], and time-frequency features [14]. A number of researchers have also applied support vector machines (SVM) for heart sound classification in recent years. The studies can also be divided according to the feature extraction methods, including wavelet [15], time, frequency and time-frequency feature-based classifiers [16]. Hidden Markov models (HMM) have also been employed for pathology classification in PCG recordings [17,18]. Clustering-based classifiers, typically the k-nearest neighbors (kNN) algorithm [19,20], have also been employed to classify pathology in PGCs. In addition, many other techniques have been applied, including threshold-based methods, decision trees [21], discriminant function analysis [22,23] and logistic regression.
Although a number of the current studies for heart sound classification are flawed because of 1) good performance on carefully-selected data, 2) lack of a separate test dataset, 3) failure to use a variety of PCG recordings, or 4) validation only on clean recordings, these methods have demonstrated potential to accurately detect pathology in PCG recordings. In this Challenge, we will focus only on the accurate classification of normal and abnormal heart sounds, especially when some heart sounds exhibit very poor signal quality. The Challenge provides the largest public collection of PCG recordings from a variety of clinical and nonclinical environments, permitting challengers to develop accurate and robust algorithms.
Quick Start
- Download the validation set and the sample MATLAB entry.
- Develop your entry by editing the existing files:
- Modify the sample entry source code file
challenge.mwith your changes and improvements. For additional information, see the Preparing an Entry for the Challenge section. - Modify the
AUTHORS.txtfile to include the names of all the team members. - Unzip
validation.zipand move thevalidationdirectory to the same directory wherechallenge.mis located. - Run your modified source code file on all the records in the training set by executing the script
generateValidationSet.m. This will also build a new version ofentry.zip. - Optional: Include a file named
DRYRUNin the top directory of your entry (where theAUTHORS.txtfile is located) if you do not wish your entry to be scored and counted against your limit. This is useful in cases where you wish to make sure that the changes made do not result in any error.
- Modify the sample entry source code file
- Submit your modified entry.zip for scoring through the PhysioNet PhysioNet/CinC Challenge 2016 project (update: submissions are now closed). The contents of entry.zip must be laid out exactly as in the sample entry. Improperly-formatted entries will not be scored.
Join our community Community Discussion Forum to get the latest challenge news, technical help, or if you would like to find partners to collaborate with.
Rules and Deadlines
Participants are asked to classify recordings as normal, abnormal (i.e. they require further evaluation by an expert for further evaluation or potential treatment) or too noisy or ambiguous to evaluate.
Entrants may have an overall total of up to 15 submitted entries over both the unofficial and official phases of the competition (see Table 1). Each participant may receive scores for up to five entries submitted during the unofficial phase and ten entries at the end of the official phase. Unused entries may not be carried over to later phases. Entries that cannot be scored (because of missing components, improper formatting, or excessive run time) are not counted against the entry limits.
All deadlines occur at noon GMT (UTC) on the dates mentioned below. If you do not know the difference between GMT and your local time, find out what it is before the deadline!
| Start at noon GMT on |
Entry limit |
End at noon GMT on |
|
| Unofficial Phase | 1 March | 5 | |
| [Hiatus] | 0 | ||
| Official Phase | 10 | 26 August |
All official entries must be received no later than the noon GMT on Friday, 26 August 2016. In the interest of fairness to all participants, late entries will not be accepted or scored. Entries that cannot be scored (because of missing components, improper formatting, or excessive run time) are not counted against the entry limits.
To be eligible for the open-source award, you must do all of the following:
- Submit at least one open-source entry that can be scored before the Phase I deadline (noon GMT on Sunday, 1 May 2016).
- Submit a draft abstract about your work on the Challenge to Computing in Cardiology no later than 14 April 2016. Please select "PhysioNet/CinC Challenge" as the topic of your abstract, so it can be identified easily by the abstract review committee.
- Submit a final abstract (about 300 words) no later than 2 May 2016. Include the overall score for at least one Phase I entry in your abstract. You will be notified if your abstract has been accepted by email from CinC during the first week in June.
- Submit a full (4-page) paper on your work on the Challenge to CinC no later than 1 September 2016.
- Attend CinC 2016 (11-14 September 2016) and present your work there.
Please do not submit analysis of this year's Challenge data to other conferences or journals until after CinC 2016 has taken place, so the competitors are able to discuss the results in a single forum. We expect a special issue from the journal Physiological Measurement to follow the conference and encourage all entrants (and those who missed the opportunity to compete or attend CinC 2016) to submit extended analyses and articles to that issue, taking into account the publications and discussions at CinC 2016.
Challenge Data
Heart sound recordings were sourced from several contributors around the world, collected at either a clinical or nonclinical environment, from both healthy subjects and pathological patients. The Challenge training set consists of five databases (A through E) containing a total of 3,126 heart sound recordings, lasting from 5 seconds to just over 120 seconds. You can browse these files, or download the entire training set as a zip archive (169 MB). Updated classification and signal quality annotations for the training set are available in the Files section under annotations/updated.
In each of the databases, each record begins with the same letter followed by a sequential, but random number. Files from the same patient are unlikely to be numerically adjacent. The training and test sets have each been divided so that they are two sets of mutually exclusive populations (i.e., no recordings from the same subject/patient were are in both training and test sets). Moreover, there are two data sets that have been placed exclusively in either the training or test databases (to ensure there are ‘novel’ recording types and to reduce overfitting on the recording methods). Both the training set and the test set may be enriched after the close of the unofficial phase. The test set is unavailable to the public and will remain private for the purpose of scoring.
Participants may note the existence of a validation dataset in the data folder. This data is a copy of 300 records from the training set, and will be used to validate entries before their evaluation on the test set. More detail will be provided in the scoring section below.
The heart sound recordings were collected from different locations on the body. The typical four locations are aortic area, pulmonic area, tricuspid area and mitral area, but could be one of nine different locations. In both training and test sets, heart sound recordings were divided into two types: normal and abnormal heart sound recordings. The normal recordings were from healthy subjects and the abnormal ones were from patients with a confirmed cardiac diagnosis. The patients suffer from a variety of illnesses (which we do not provide on a case-by-case basis), but typically they are heart valve defects and coronary artery disease patients. Heart valve defects include mitral valve prolapse, mitral regurgitation, aortic stenosis and valvular surgery. All the recordings from the patients were generally labeled as abnormal. We do not provide more specific classification for these abnormal recordings. Please note that both training and test sets are unbalanced, i.e., the number of normal recordings does not equal that of abnormal recordings. You will have to consider this when you train and test your algorithms.
Both healthy subjects and pathological patients include both children and adults. Each subject/patient may have contributed between one and six heart sound recordings. The recordings last from several seconds to up to more than one hundred seconds. All recordings have been resampled to 2,000 Hz and have been provided as .wav format. Each recording contains only one PCG lead.
Please note that due to the uncontrolled environment of the recordings, many recordings are corrupted by various noise sources, such as talking, stethoscope motion, breathing and intestinal sounds. Some recordings were difficult or even impossible to classify as normal or abnormal. Therefore we have given the challengers the choice to classify some recordings as ‘unsure’ and we penalize this in a different manner. Therefore, your classifications for the heart sound recordings could be three types: normal, abnormal and unsure (too noisy to know). The detailed scoring mechanism could be found in Scoring section.
Note: A paper to provide a detailed description of all the heart sound data in PhysioNet/CinC Challenge 2016 is expected to appear in the Journal Physiological Measurement on or about July 2016. We will post a preprint of it on this site soon to help you understand the Challenge more thoroughly and may help in improving your submitted algorithms in the Official Phase.
Sample Submission
As a starting point, we have provided an example entry (sample2016.zip), implemented using Matlab, which provides state of the art segmentation and rudimentary classification. This code first segments the heart sounds using Springer’s improved version of Schmidt’s method [5,9], which uses a Hidden Markov Model (HMM) that has been trained (using database ‘a’ of the training set) to identify four ‘states’; S1, S2, systole and diastole. Thereafter, 20 features are extracted from the timings of the states and a logistic regression classifier (again, trained on database ‘a’ of the training data) provides the classification of the recording as normal or abnormal. For more information about this algorithm, see the released Logistic Regression-HSMM-based Heart Sound Segmentation software package on PhysioToolkit. For the segmentation annotations for the training set from Springer’s segmentation algorithm, see the annotations/springer_alg folder in the Files section. For hand-corrected segmentation annotations for the training set, see the annotations/hand_corrected folder and Liu et al., "An open access database for the evaluation of heart sound algorithms", Physiol. Meas, 2016 Dec;37(12):2181-2213. doi: 10.1088/0967-3334/37/12/2181 for details.
A simpler version of this code (sample2016b.zip), using Schmidt's original algorithm, is faster and works in GNU Octave as well as in Matlab.
You may want to begin with this framework, and add more intelligent approaches, or discard it completely and start from scratch. The features and classifier are not necessarily recommended and they are only provided as an example benchmark approach. The beat segmentation algorithm is, however, state of the art. We therefore suggest you concentrate on adapting this to provide better features. Note also that we have not optimized the training of either the HMM, the features chosen or selected, the splitting of the data, or the classifier. We suggest you consider these issues carefully.
Preparing an entry for the challenge
To participate in the challenge, you will need to create software that is able to read the test data and output the final classification result without user interaction in our test environment. A sample entry (sample2016.zip), written in MATLAB, is available to help you get started. In addition to MATLAB, you may use any programming language (or combination of languages) supported using open-source compilers or interpreters on GNU/Linux, including C, C++, Fortran, Haskell, Java, Octave, Perl, Python, and R.
If your entry requires software that is not installed in our sandbox environment, please let us know before the end of Phase I. We will not modify the test environment after the start of Phase II of the challenge.
Your entry must be in the format of a zip or tar.gz archive, containing the following files:
setup.sh, abashscript run once before any other code from the entry; use this to compile your code as needednext.sh, abashscript run once per training or test record; it should analyze the record using your code, writing the results to the fileanswers.txt.answers.txt, a text file containing the results of running your program on each record in the validation set. These results are used for checking that your program is working correctly, not for ranking entries (see below).AUTHORS.txt, a plain text file listing the members of your team who contributed to your code, and their affiliations.LICENSE.txt, a text file containing the license for your software. The sample entry is licensed under the GNU GPL. All entries are assumed to be open source and will eventually be released on PhysioNet.
See the comments in the sample entry's setup.sh and next.sh if you wish to learn how to customize these scripts for your entry.
We verify that your code is working as you intended, by running it on the validation set, which consists of approximately 10% of the training set. We then compare the answers produced by your code with the contents of the answers.txt file that you submit as part of your entry. Using a small portion of the training set means you will know whether your code passed or failed to run in approximately an hour or less. If your code passes this validation test, it is then evaluated and scored using an approximately representative 20% of the hidden test set. By selecting a random 20% subset of the test set, not only do you receive your score in a more timely manner, but it also prevents you from over-fitting on the test data through multiple entries. Towards the end of the official phase we will run your code on increasingly larger portions of the test set. The score on the complete test set determines the ranking of the entries and the final outcome of the Challenge.
In addition to the required components, your entry may include a file named DRYRUN. If this file is present, your entry is not evaluated using the hidden test data, and it will not be counted against your limit of entries per phase; you will receive either a confirmation of success or a diagnostic report, but no scores. Use this feature to verify that none of the required components are missing, that your setup.sh script works in the test environment, and that your next.sh script produces the expected output for the training data within the time limits.
Scoring
If your entry is properly formatted, and nothing is missing, it is tested and scored automatically, and you will receive your provisional scores when the test is complete (this will take several hours, depending on how complex your entry is). If you receive an error message instead, read it carefully and correct the problem(s) before resubmitting.
The overall score for your entry is computed based on the number of records classified as normal, uncertain, or abnormal, in each of the reference categories. These numbers are denoted by Nnk, Nqk, Nak, Ank, Aqk, Aak, as follows:
| Entry's output | ||||
| Normal (-1) | Uncertain (0) | Abnormal (1) | ||
| Reference label | Normal, clean | Nn1 | Nq1 | Na1 |
| Normal, noisy | Nn2 | Nq2 | Na2 | |
| Abnormal, clean | An1 | Aq1 | Aa1 | |
| Abnormal, noisy | An2 | Aq2 | Aa2 | |
Weights for the various categories are defined as follows (based on the distribution of the complete test set):
The modified sensitivity and specificity are defined (based on a subset of the test set):
The overall score is then the average of these two values, .
Obtaining complimentary MATLAB licenses
The MathWorks has kindly decided to sponsor Physionet's 2016 Challenge both through additional prize money for the winners and through complimentary licenses for Challenge participants for the duration of the Challenge. User can apply for a license and learn more about MATLAB support through The MathWorks' PhysioNet Challenge link. If you have questions or need technical support, please contact The MathWorks at academicsupport@mathworks.com.
Challenge Results
Listed below are the top-scoring programs submitted in the PhysioNet/Computing in Cardiology Challenge 2016. Please refer to the AUTHORS.txt and LICENSE.txt file included with each entry for information about attribution and licensing. For more information about the details of these algorithms, see the corresponding papers.
Top-Scoring 2016 Challenge Entries
Papers
The following paper is an introduction to the challenge topic, with a summary of the challenge results and a discussion of their implications. Please cite this publication when referencing the Challenge.
Classification of Normal/Abnormal Heart Sound Recordings: the PhysioNet/Computing in Cardiology Challenge 2016
Gari D. Clifford, Chengyu Liu, Benjamin Moody, David Springer, Ikaro Silva, Qiao Li, Roger G. Mark
Over 12 papers were presented at Computing in Cardiology 2016. These papers have been made available by their authors under the terms of the Creative Commons Attribution License 3.0 (CCAL). They are available here. We wish to thank all of the authors for their contributions.
Access
Access Policy:
Anyone can access the files, as long as they conform to the terms of the specified license.
License (for files):
Open Data Commons Attribution License v1.0
Discovery
Topics:
sound
heart
challenge
phonocardiogram
Corresponding Author
Files
Total uncompressed size: 1.1 GB.
Access the files
- Download the ZIP file (1011.4 MB)
- Access the files using the Google Cloud Storage Browser here. Login with a Google account is required.
-
Access the data using the Google Cloud command line tools (please refer to the gsutil
documentation for guidance):
gsutil -m -u YOUR_PROJECT_ID cp -r gs://challenge-2016-1.0.0.physionet.org DESTINATION
-
Download the files using your terminal:
wget -r -N -c -np https://physionet.org/files/challenge-2016/1.0.0/
-
Download the files using AWS command line tools:
aws s3 sync s3://physionet-open/challenge-2016/1.0.0/ DESTINATION
| Name | Size | Modified |
|---|---|---|
| annotations | ||
| papers | ||
| sources | ||
| training-a | ||
| training-b | ||
| training-c | ||
| training-d | ||
| training-e | ||
| training-f | ||
| validation | ||
| annotations.zip (download) | 10.1 MB | 2022-07-28 |
| figure1.png (download) | 99.9 KB | 2019-04-17 |
| mathworks.png (download) | 14.4 KB | 2019-04-17 |
| sample2016.zip (download) | 50.4 KB | 2019-04-17 |
| sample2016b.zip (download) | 1.1 MB | 2019-04-17 |
| training.zip (download) | 181.3 MB | 2016-05-18 |
| validation.zip (download) | 17.5 MB | 2016-03-18 |